Investigating the transport pathway of Shiga toxin to add a new dimension to treatment strategies
With the spread of COVID-19, research on infectious diseases is now garnering more public attention than ever before. Research to elucidate the toxicity expression mechanisms of infectious agents for therapeutic drug development is constantly advancing. Assistant Professor Miho Takahashi from the Department of Medical Life Systems, who specializes in clinical pharmacology, is utilizing the accumulating research to tackle this formidable challenge.
Enterohemorrhagic Escherichia coli (E. coli) infections, such as that of O-157, are known to cause diarrhea with abdominal pain and bleeding, and in some cases, impair brain and renal functions leading to severe complications. In Japan, more than 3,000 people contract this disease each year, out of whom approximately 6 to 7% develop severe symptoms. According to reports, some cases even lead to death. This powerful toxicity comes from a bacterial toxin known as Shiga toxin (Stx).
“There are two kinds of Shiga toxin, Stx1, and Stx2. While their structures are quite similar, their toxicity is clearly different. While it takes a few hundred nanograms of Stx1 for the toxin to be lethal for a mouse, it only takes one nanogram of Stx2 to achieve the same. Clearly, Stx2 has higher toxicity. In clinical practice, it is said that those who were infected by bacteria that produce Stx2 tend to develop more severe conditions. It was not known why only Stx2 exhibits high toxicity. Therefore, I wanted to discover this high toxicity expression mechanism and make it a pathway for developing therapeutic drugs,” said Dr. Takahashi.
Shiga toxin’s structure consists of one molecule A-subunit—the toxin body—and five molecule B-subunits—the transporters. When Stx enters the human cells, the B-subunit pentamer targets the glycolipid on the cell’s surface called Gb3. It passes through the Golgi in the cell, and is ultimately carried to the endoplasmic reticulum, while only the A-subunit is released into the cytoplasm. Subsequently, the A-subunit deactivates the ribosome—where protein synthesis occurs—and inhibits the synthesis of the proteins necessary for the cells.
Dr. Takahashi focused on the recycling pathway, which is the route taken by Shiga toxin to exit the cell after entering it (Figure 1). “When the amount of Stx1 and Stx2 released from the cells were compared, it was found that three to four more amounts of Stx2 had been released compared to that of Stx1. When I investigated the reason for this difference, I realized there was a difference in how Stx1 and Stx2 moved within a cell. Stx1 heads towards the endoplasmic reticulum via the early endosome. On the other hand, Stx2 may take routes similar to those taken by Stx1 on occasion, but it was found that Stx2 has a much easier time getting on to the recycling pathway than Stx1. Stx2 traveled on the recycling pathway and was actively being released. Furthermore, when I investigated the released Stx2, I found two types, those which traveled on an exosome (Exo-Stx2) and those which did not (free-Stx2 ).”
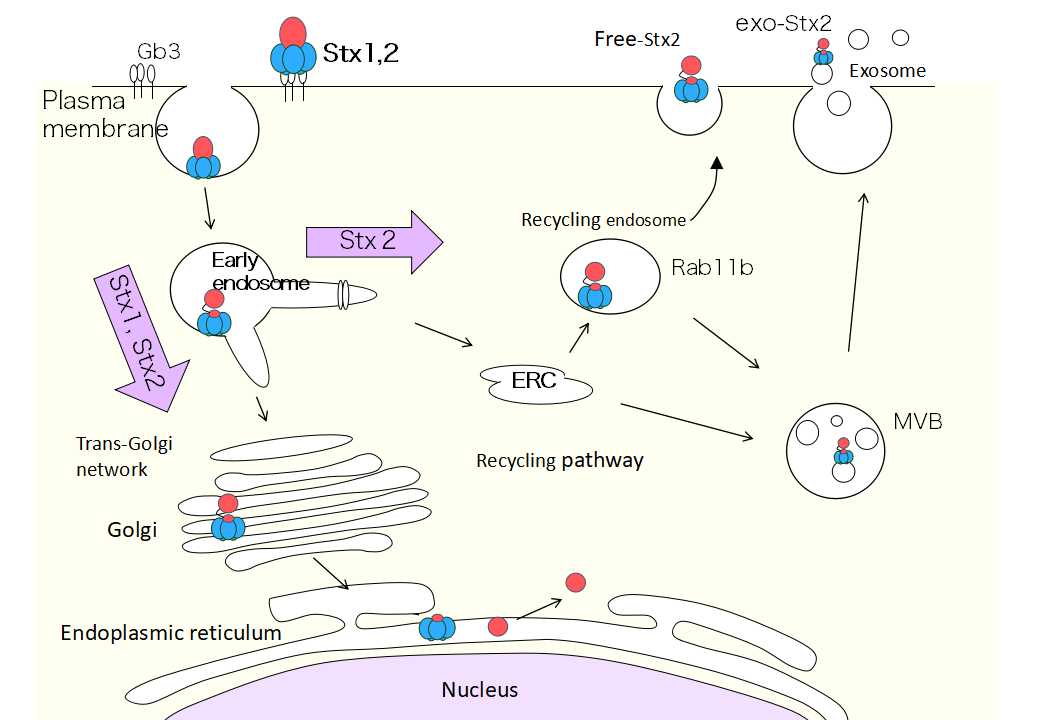
(Figure 1) Stx2 released from the cell via the recycling pathway
When the Exo-Stx2 and the free-Stx2 were added to Vero cells, Dr. Takahashi found no difference in cytotoxicity between the two; but when it came to individual mice, she found that Exo-Stx2 expressed greater toxicity than free-Stx2. (Figure 2) shows the comparison of the survival rate of mice after the intravenous administration of either Exo-Stx2 or free-Stx2. At all concentrations, Exo-Stx2 shows a higher lethality.
Furthermore, Dr. Takahashi noticed during the experiment that mice that had been administered Exo-Stx2 produced a significantly greater amount of urine than normal. When the renal tissue of the Exo-Stx2-administered mice was compared to the free-Stx2-administered mice, she found the tubular epithelial cells of Exo-Stx2 administered mice were severely damaged and shedding. From this, it became clear that the reabsorption function of the tubular cells was not active and thus led to an increase in the urine volume.
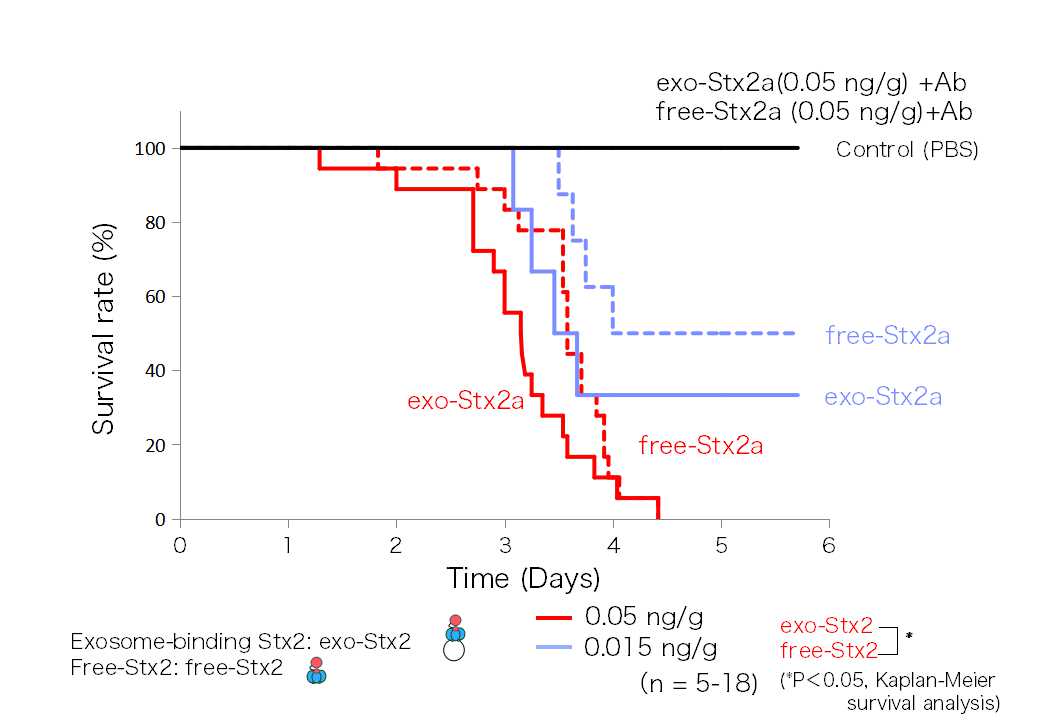
(Figure 2) Exo-Stx2 shows high toxicity towards mice
“There are still so many unanswered questions, such as ‘What would cause the tubular epithelial cells to become severely damaged and shed with Exo-Stx2?’ I wish to continue my research to find these answers,” said Dr. Takahashi.
The research that began with comparing the differences between the structure and toxicity of Shiga toxin’s Stx1 and Stx2 led to the discovery of the existence of Exo-Stx2 and its high toxicity. Further elucidation on the mechanism of Exo-Stx2 toxicity expression may lead to new treatment strategies in the future.
Creating new inhibitors with the highly practical multivalent peptide library technique
Parallel to her research on the development of inhibitors by elucidating the toxicity expression mechanism, Dr. Takahashi is also working on drug discovery using a peptide library, a mixture of peptides through a random combination of amino acids. The multivalent peptide library technique (Figure 3) developed by Professor Kiyotaka Nishikawa, who leads the Molecular Biochemistry Laboratory that Dr. Takahashi belongs to, is an innovative drug discovery platform that enables the acquisition of motifs that can inhibit the interactions between multivalent molecules. Using this highly versatile method that allows ease of usage for peptide synthesis, she is conducting research on various molecules to target the Shiga toxin, cholera toxin, and BCR-ABL, which causes chronic myeloid leukemia, and others to create peptide molecules that will become candidates for pharmaceutical products.
The pharmaceutical industry is paying a lot of attention to peptide-based drug development, which has greater potential than conventional pharmaceutical molecules. “We are actively conducting joint research with other research institutions in the academia. Going forward, I would like to collaborate with the industry too,” says Dr. Takahashi.
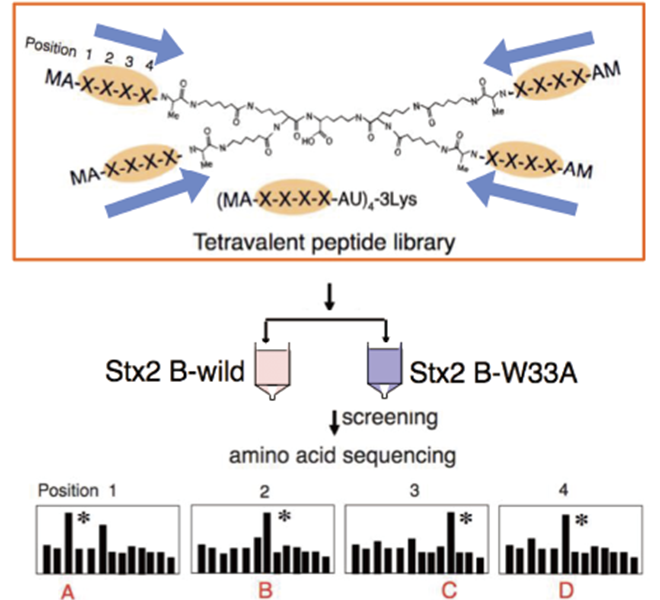
(Figure 3) Multivalent peptide library technique
A technique in which target-specific control molecules are obtained from a mixture of multivalent peptides (random peptide library) having XXXX (random amino acid sequences) based on their affinity to the target.
Rising to the challenge of developing COVID-19 therapeutic agents via established methods
Dr. Takahashi is also using the multivalent peptide library technique in her project selected as a COVID-19 Research Project titled “The development of COVID-19 therapeutic agents targeting the receptor-binding domain of the novel coronavirus (SARS-CoV-2) spike protein” to find clues for drug discovery. “There is a spike (S) protein with a trimer structure on the surface of the novel coronavirus. We know that the virus penetrates the cells when the S protein binds to the ACE2 protein on human cell membranes. I am searching for the possibility of developing a therapeutic agent by creating a motif that would prevent that binding,” Dr. Takahashi explains.
While Dr. Takahashi has found much success through years of experimentations, she admits it often does not go the way she had hoped. “Even if the results seem negative, they may provide hints to new discoveries. I hope not to miss any factors and consider all possibilities.” Even though she spoke with calm tones, her insatiable passion for research can be heard loud and clear.
